Balancing the Scales: Post-Transcriptional Dosage Compensation in Chickens and Platypuses

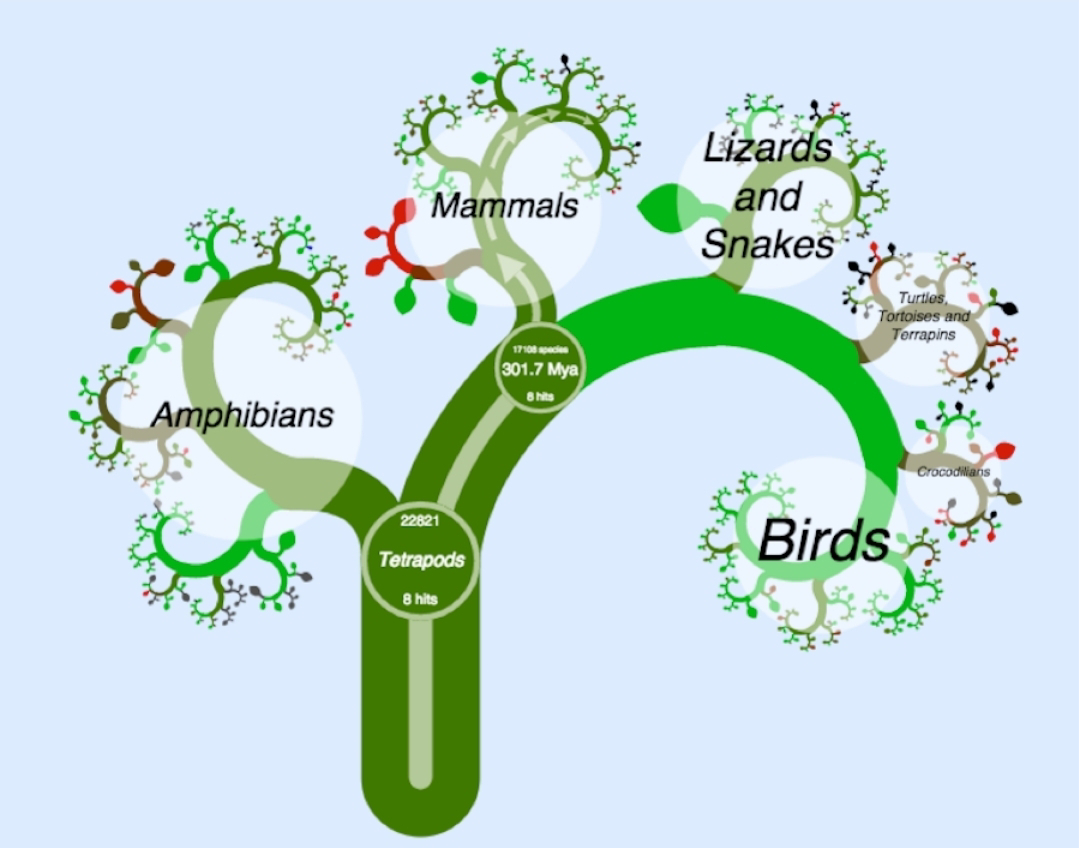
The intricate dance of gene expression on sex chromosomes has long fascinated scientists. Sex chromosomes, such as the XY system in mammals or the ZW system in birds, often present a challenge: how to ensure that genes on these chromosomes are expressed at appropriate levels in both sexes, despite differences in chromosome number. The prevailing model, inspired by X-chromosome inactivation in mammals, suggested that dosage compensation – mechanisms to equalize gene expression between sexes – was a universal necessity in vertebrates. However, recent findings have challenged this view. The Study In the groundbreaking journal article “Incomplete transcriptional dosage compensation of chicken and platypus sex chromosomes is balanced by post-transcriptional compensation”, researchers Lister et al. (2024) provide compelling evidence for a novel model of dosage compensation. They focused on two evolutionarily distant species – the chicken (ZW system) and the platypus (multiple X ...