The Regulatory Symphony: How Epigenetics and Noncoding RNA Orchestrate Human Brain Evolution Beyond Neo-Darwinism
The development of the human brain, with its unparalleled cognitive capabilities, stands as one of the most profound transformations in life's history. For decades, the dominant explanatory framework, rooted in neo-Darwinism (the Modern Synthesis), focused primarily on natural selection acting upon random mutations within protein-coding genes. This view posits that gradual changes in the amino acid sequences of proteins conferred survival advantages, leading over millennia to the complex structures and functions underlying human thought. However, accumulating evidence, particularly concerning regulatory DNA and RNA, suggests this gene-centric view is incomplete. The emerging picture reveals that the evolution of how, when, and where genes are expressed—governed by regulatory DNA, epigenetics, and noncoding RNAs (ncRNAs)—played a dominant, role in shaping the human brain, presenting significant challenge to the traditional neo-Darwinian narrative.
Central to this expanded view is the realization that organismal complexity, particularly brain complexity, doesn't correlate neatly with the number of protein-coding genes. Humans possess roughly the same number of such genes as far simpler organisms. The difference, it appears, lies significantly in the regulatory landscape. Vast stretches of the genome, once dismissed as "junk DNA," are now recognized as containing crucial regulatory elements like enhancers, silencers, and promoters.
Mutations in these regions don't alter the protein product itself but can dramatically change the gene's expression pattern—its timing during development, its location within specific brain regions, and its responsiveness to environmental cues. Rapid development of human-specific enhancers associated with genes involved in neural development points towards regulatory changes being a key engine driving cognitive divergence.Epigenetics: Tuning Gene Expression in Development and Evolution:
Epigenetics adds another layer of regulatory complexity. Mechanisms like DNA methylation, histone modifications (acetylation, methylation, phosphorylation), and chromatin remodeling act as molecular switches or dials, influencing gene accessibility and expression levels without altering the underlying DNA sequence.
These epigenetic marks are crucial for normal brain development, orchestrating cell differentiation, synaptic plasticity, and cognitive function.From an evolutionary perspective, epigenetics introduces intriguing possibilities. While traditionally viewed as resetting each generation, evidence suggests that epigenetic marks can be influenced by environmental factors (diet, stress, toxins) and, controversially, exhibit transgenerational inheritance.
Even without direct inheritance, epigenetic plasticity allows organisms to fine-tune gene expression in response to their environment within a lifetime.This capacity for environmentally responsive gene regulation, particularly during critical developmental windows, could influence the pressures acting on underlying genetic variation. If epigenetic states bias developmental trajectories or physiological responses in ways that enhance survival or reproduction, they can indirectly shape the course of adaptation. This offers a mechanism for potentially faster adaptation or diversification compared to relying solely on random DNA sequence mutations. It introduces a nuance where the environment doesn't just select among random variations but can also influence the phenotypic expression upon which selection acts, blurring the strict separation of inheritance and environment central to classical neo-Darwinism.Noncoding RNA: The Dark Matter of the Genome Takes Center Stage:
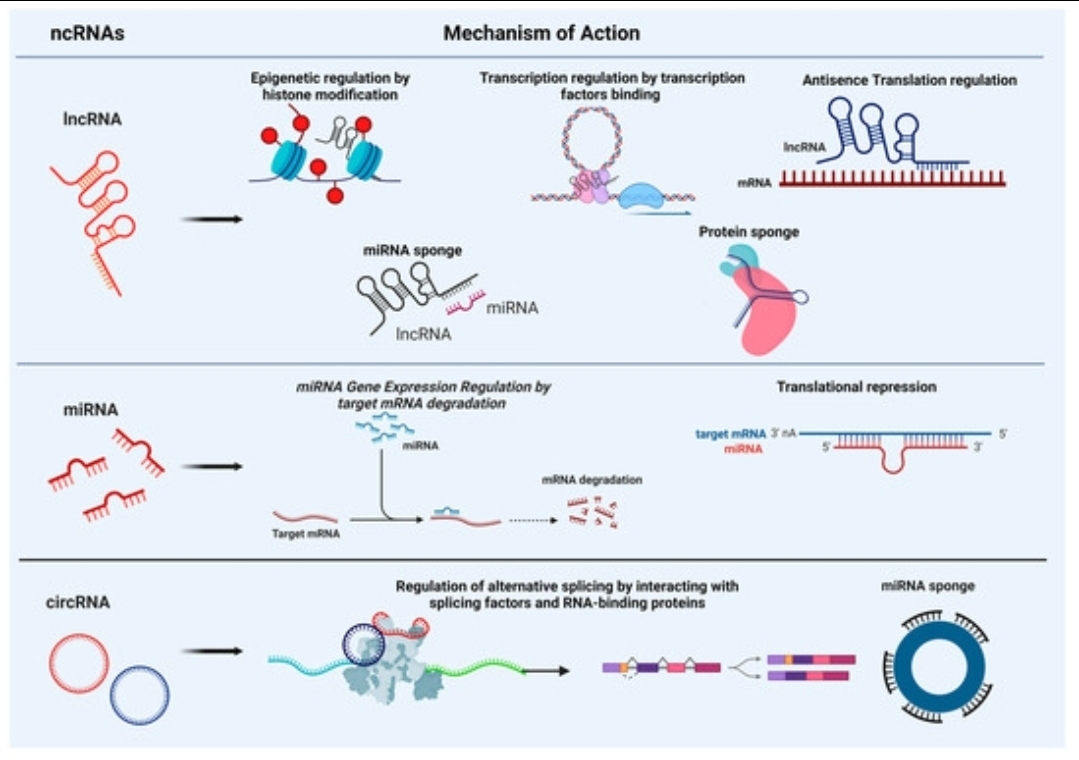
Perhaps one of the most dramatic shifts in understanding genome function comes from the burgeoning field of noncoding RNA (ncRNA). Far from being mere intermediaries in protein synthesis, ncRNAs encompass a diverse repertoire of molecules (including microRNAs, long noncoding RNAs, circular RNAs) with profound regulatory roles.
They operate at multiple levels: degrading messenger RNAs (mRNAs), inhibiting translation, modifying chromatin structure, scaffolding protein complexes, and influencing alternative splicing.The brain is particularly rich in diverse and dynamically expressed ncRNAs.
Specific microRNAs fine-tune synaptic function and neuronal development. Long noncoding RNAs (lncRNAs) are increasingly implicated in establishing and maintaining cell identity in the nervous system and regulating complex gene networks involved in cognition. Many ncRNAs show human-specific expression patterns or sequences, suggesting they have been targets of recent adaptation. Because ncRNAs can regulate entire networks of genes simultaneously, subtle changes in their sequence, structure, or expression can cascade into significant alterations in cellular function and, ultimately, brain organization and capability. Their development provides a powerful mechanism for generating phenotypic novelty and complexity without necessarily inventing entirely new protein-coding genes.The focus is now on regulatory elements, epigenetics, and ncRNAs rather than neo-Darwinism:
Regulatory Variation as a Primary Driver: Evolution can proceed rapidly by altering gene regulation rather than solely protein structure. This provides a more plausible mechanism for large phenotypic changes, like those seen in brain evolution, without requiring vast numbers of coding sequence mutations.
Environmental Influence on Variation: Epigenetic mechanisms introduce a more direct role for the environment in shaping the expressed phenotype upon which selection acts, adding a Lamarckian flavor (though the mechanisms differ from classical Lamarckism) that complicates the strict genotype-phenotype map.
Network-Level Changes: ncRNAs demonstrate how evolution can act on regulatory networks, allowing for coordinated changes across many genes simultaneously, potentially leading to more integrated and complex adaptations.
In essence, the story of human brain evolution is becoming less about just changing the protein "bricks" and more about evolving the complex "blueprint" and "control systems"—the regulatory DNA, epigenetic modifications, and ncRNA networks—that dictate how those bricks are assembled. This regulatory perspective challenges the core principles of mutation and selection showing change operates on a much richer, more dynamic, and interconnected genomic landscape than previously appreciated. Understanding these regulatory layers is crucial for deciphering the intricate evolutionary path that led to the human mind and for formulating a more comprehensive evolutionary synthesis for the 21st century.










Comments
Post a Comment