The Flexible Framework: How Intrinsically Disordered Proteins Challenge Neo-Darwinian Orthodoxy
The established dogma of molecular biology, a cornerstone of neo-Darwinian thought, has long been anchored to a rigid principle: a protein's function is dictated by its precise, three-dimensional structure.
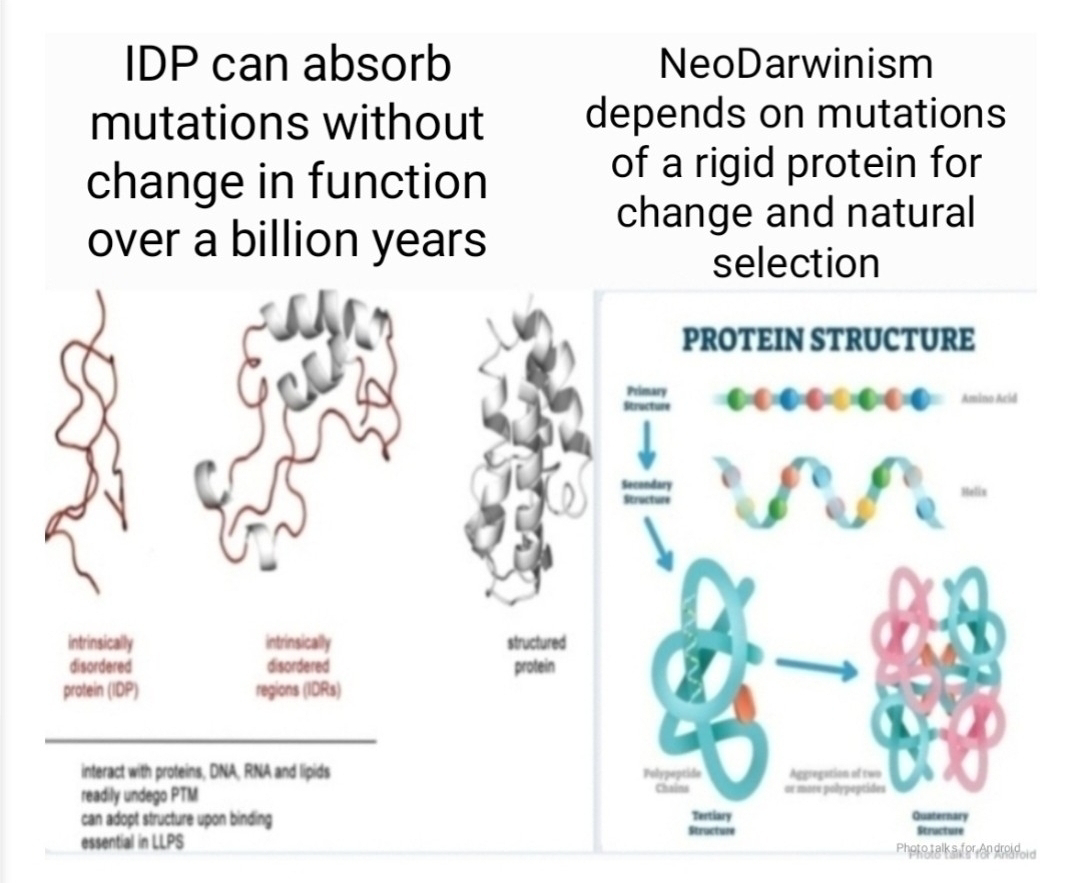
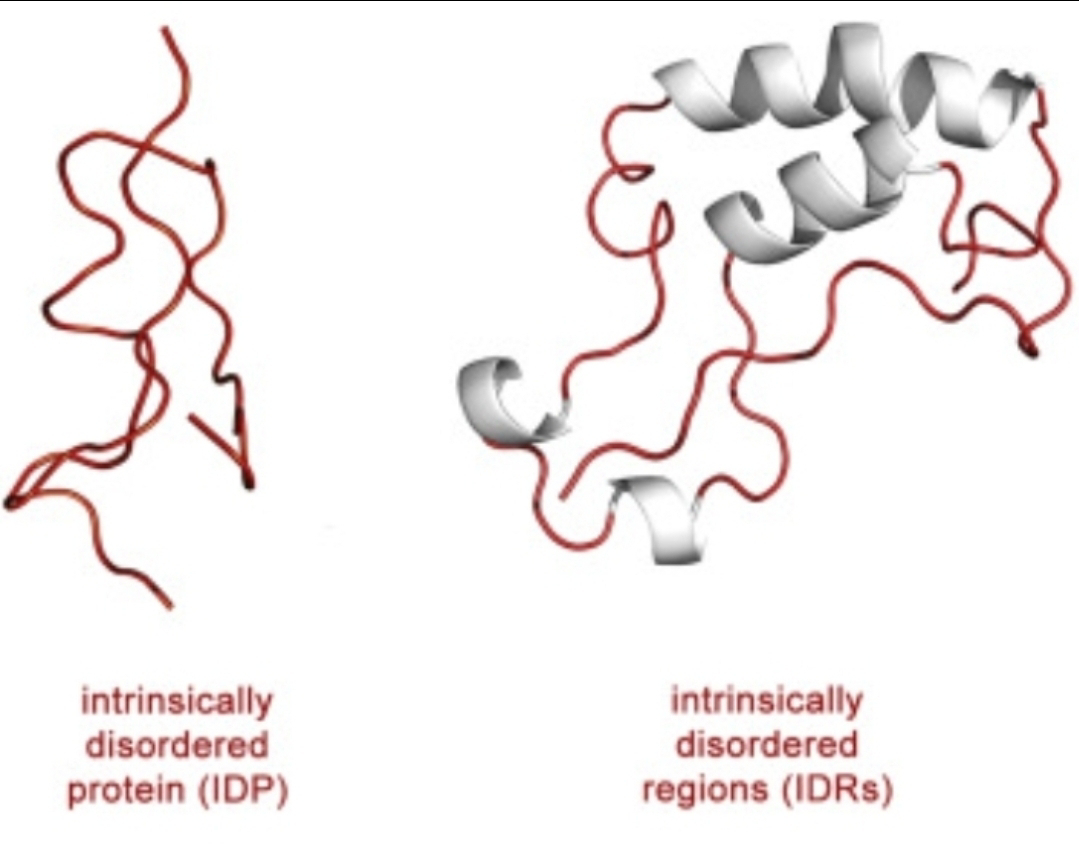
This "structure-function" paradigm suggests a straightforward evolutionary pathway where random genetic mutations lead to changes in protein structure, which are then selected for or against based on their functional consequences. However, the discovery and burgeoning understanding of intrinsically disordered proteins (IDPs) are introducing a profound plot twist to this narrative. These proteins, which lack a stable, folded structure, are not evolutionary anomalies but are abundant and crucial players in cellular life, presenting a significant conceptual challenge to the strictly defined tenets of neo-Darwinism.At its core, neo-Darwinism posits that evolution proceeds through the gradual accumulation of random mutations within a population. These mutations, it is argued, lead to phenotypic variation, and natural selection then acts upon this variation, favoring traits that enhance survival and reproduction. This model implicitly relies on a relatively direct and predictable relationship between genotype and phenotype. A change in a gene should, in theory, lead to a specific change in a protein's structure, altering its function in a discernible way that can be tested by the pressures of the environment.The journal article "Introduction to Intrinsically Disordered Proteins (IDPs)" and subsequent research in the field fundamentally disrupt this linear, structure-centric view. IDPs exist not as single, static conformations, but as dynamic ensembles of interconverting structures. Their functional versatility stems from this very lack of a fixed shape. This inherent plasticity allows a single IDP to bind to multiple partners, act as a hub in signaling networks, and respond dynamically to changes in the cellular environment. This multi-functionality and environmental sensitivity challenge the neo-Darwinian framework in several key ways.
Firstly, IDPs complicate the direct line from genotype to phenotype. A mutation in the gene encoding an IDP might not lead to a simple, predictable change in function. Instead, it could alter the ensemble of conformations, subtly shifting the protein's interaction landscape and its responsiveness to cellular signals. This introduces a layer of complexity and context-dependence that is not easily captured by traditional models of gene-centric selection. The phenotype is no longer a static target for selection but a dynamic potentiality, realized through the interplay of the IDP's sequence and its environment.
Secondly, the adaptability of IDPs suggests a mechanism for rapid evolutionary innovation that goes beyond the slow, incremental changes envisioned by classical neo-Darwinism. Because a single IDP can perform multiple functions, a mutation that modifies one of its interactions might not completely abrogate its other essential roles. This functional "scaffolding" allows for greater evolutionary experimentation. An organism with a rich complement of IDPs may be more "evolvable," capable of exploring new phenotypic space without the potentially catastrophic consequences of altering a rigidly structured, single-function protein. This inherent robustness and potential for rapid adaptation challenge the notion that evolution must always proceed through small, gradual steps.
Furthermore, the discovery of IDPs has profound implications for our understanding of the origins of biological complexity. The intricate signaling and regulatory networks that characterize eukaryotic life are heavily reliant on the functional promiscuity of IDPs. Their ability to form transient, context-dependent interactions is essential for the complex choreography of cellular processes. A neo-Darwinian explanation for the evolution of such complexity would typically involve a long and arduous process of gene duplication and divergence, with each new function arising from a specific set of mutations. The existence of IDPs suggests a more fluid and efficient route to complexity. A single, disordered protein can act as a versatile building block, its functions expanding and diversifying not just through genetic changes but also through alterations in its cellular environment and interacting partners. This provides a more plausible mechanism for the emergence of the complex regulatory systems that are a hallmark of higher organisms.
In essence, the study of intrinsically disordered proteins challenges the core principles of mutation and natural selection. It enriches and refines our understanding of how these processes operate. The "Introduction to Intrinsically Disordered Proteins (IDPs)" and the wealth of research that has followed have unveiled a world of biological organization that is far more dynamic and less deterministic than previously imagined. By demonstrating that function can arise from controlled disorder and that a single protein can embody a multitude of possibilities, IDPs challenge the rigid, structure-centric view of the cell that has long underpinned neo-Darwinian thought. They suggest that the evolutionary dance is not a stately waltz of gradual changes but a dynamic and improvisational performance, where the flexibility of its molecular players is just as important as the precision of their steps. This more nuanced perspective opens up exciting new avenues for understanding the evolution of life in all its complexity.




Comments
Post a Comment